Ph.D. - Statistical Physics
Rutgers University - 1987

Michael Zhang
Cecil H. and Ida Green Distinguished Chair of Systems Biology Science
Professor - Biological Sciences, Also affiliated with Bioengineering and Mathematics
Director of Center for Systems Biology
B.S. in Mech Eng. from USTC in 1981 and Ph.D. in Stat Phys from Rutgers U. in 1987. After 3 yr postdoctoral research in Applied Math at Courant Inst. NYU, moved to CSHL from Genome Res Fellow to full professor until moved to UTD in 2009.
Professional Preparation
B.S. - Mechanical Engineering
University of Science and Technology of China - 1981
University of Science and Technology of China - 1981
Research Areas
Research Interests
The long-term goal of research in our lab is to use mathematical and statistical methods to identify functional elements in eucaryotic genomes, especially the genes and their control and regulatory elements. A genome is the program book of a life, genome research will lead to eventual decoding of the entire genetic language of life and its grammar. Driven by the Human Genome Project, our current interest is on two related problems: genome/chromatin organization and gene regulation networks. At the transcriptional level, identification of cis-elements (both genetic and epigenetic) is the key focus. We are increasingly interested post-transcriptional regulations, especially at splicing regulation and translational regulation. Constitutive coding exons are relatively easy to identify, the greatest challenge lies in the identification of end exons and alternatively spliced exons that are often tissue- and developmental specific. Since this requires the study of many important control and regulatory elements for gene expression, this link between gene structure and function at the genomic or pre-/pri- RNA level requires high-throughput functional studies. Detecting cis regulatory elements and modeling gene expression networks are difficult challenges in the functional genomics era. Working closely with bench-scientists, our investigation will undoubtedly contribute to the understanding of genome organization as well as gene expression and regulation mechanisms, which will in turn have a profound impact on biology and medicine.Publications
Tang M.H., Varadan V., Kamalakaran S., Zhang M.Q., Dimitrova N., Hicks J. (2013) Major chromosomal breakpoint intervals in breast cancer co-localize with differentially methylated regions. Front Oncol. 2:197. PMID:23293768 2013 - Publication
Akerman M., Zhang M.Q., Krainer A. (2013) ProteoMirExpress: inferring microRNA-centered regulatory networks from high throughput proteomic and mRNA expression data. MCP, Submitted. 2013 - Publication
Megha Rajaram; Jianping Zhang; Tim Wang; Cem Kuscu; Mamoru Kato; Vladimir Grubor; Robert Weil; Asluag Helland; Anne-Lise Borrenson-Dale; Kathleen Cho; Douglas A Levine; Alan N Houghton; Jedd D Wolchok; Lois Myeroff; Sanford D Markowitz; Michael Zhang; Alex Krasnitz; Robert Lucito; David Mu; Scott Powers (2013) Two Distinct Categories of Focal Deletions in Cancer Genomes. PLoS ONE, submitted. 2013 - Publication
Wen-Yu Chung, Robert J. Schmitz, Tanya Biorac, Delia Ye, Miroslav Dudas, Gavin D. Meredith, Christopher C. Adams, Joseph R. Ecker and Michael Q. Zhang (2013) Constructing hepitypes: phasing local genotype and DNA methylation. JNSNE, In press. 2013 - Publication
Xie W., Schultz M., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J.W., Tiam S., Hawkins D.R., Leung D., Yang H., Wang T., Swanson S.A., Zhang J., Zhu Y., Lee A.Y., Kim A., Nery J., Urich M.A., Kuan S., Yen C., Klugman S., Yu P., Suknuntha K., Propson N.E., Chen H., Edsall L.E., Wagner U., Li Y., Ye Z., Kulkarni A., Xuan Z., Chung W., Chi N.C., Antosiewicz-Bourget J., Slukvin I., Stewart R., Zhang M.Q., Wang W., Thomson J.A., Eckert J.R., Ren B. (2013) Epigenomic Analysis of Multi-lineage Differentiation of Human Embryonic Stem Cells. Cell, In press. 2013 - Publication
Xiaotu Ma, Ashwinikumar Kulkarni, Zhihua Zhang, Zhenyu Xuan, Robert Serfling and Michael Q. Zhang. A highly efficient and effective motif discovery method for ChIP-seq/ChIP-chip data using positional information. Nucleic Acids Research, 2012, 1–1. 2012 - Publication
Zhang M Q. Dissecting Splicing Regulatory Network by Integrative Analysis of CLIP-Seq Data. Chapter in Bioinformatics for High Throughput Sequencing. Rodríguez-Ezpeleta, Naiara; Hackenberg, Michael; Aransay, Ana M. (Eds.) 2012, p209. 2012 - Publication
Appointments
Cecil H. and Ida Green Distinguished Chair Professor in Systems Biology
UT Dallas [2009–2029]
UT Dallas [2009–2029]
Additional Information
Molecular Biology and Bioinformatics Training
- Life Technology Training Center, Recombinant DNA Techniques I and II, (Lab.Course Certificate), Mar. and Sept., 1992.
- SIAM Computational Biology Workshop (Training Certificate), Rutgers University, Aug., 1992.
- Advanced Automated/Diagnostic DNA Sequencing Workshop, Baylor College of Medicine, Jun., 1993.
- Molecular Genetics, Cell Biology and Cell Cycle of Fission Yeast Course, CSHL, Nov., 1993.
- Experimenting on Differential Display (Liang and Pardee,1992) Method by Internal Dye-labeling in David Beach's Lab., 5 months,1994.
- Advanced Genome Sequence Analysis Course, CSHL, Mar., 1995.
- Nucleic Acid & Protein Sequence Analysis Workshop, Pittsburgh Supercomputing Center, Jun. 1995.
- Unix (Solaris 2.5x) system administration certificate from Sun microsystems, 1997.
News Articles
Scientists Advance Understanding of DNA Responsible for Diseases
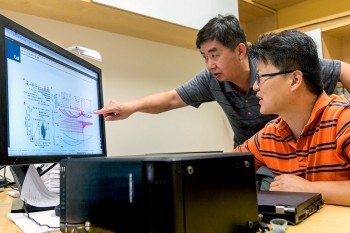 Researchers from the School of Natural Sciences and Mathematics at The University of Texas at Dallas have teamed with colleagues at the UT Southwestern Children’s Medical Center Research Institute to create a new method for understanding the causes of genetic diseases, such as sickle cell anemia and some cancers.
Researchers from the School of Natural Sciences and Mathematics at The University of Texas at Dallas have teamed with colleagues at the UT Southwestern Children’s Medical Center Research Institute to create a new method for understanding the causes of genetic diseases, such as sickle cell anemia and some cancers. The new system allows researchers to define the molecular structures that control the activity of regulatory DNA sequences in the human genome.
Dr. Michael Q. Zhang and postdoctoral researcher Dr. Yong Chen from Zhang’s Computational Biology Laboratory — along with assistant professor of biological sciences Dr. Zhenyu Xuan — collaborated on the formulation of the system called CAPTURE, or CRISPR Affinity Purification in situ of Regulatory Elements. Their work was published in a recent issue of the journal Cell.
Scientist Flourishes in Eugene McDermott Graduate Fellows Program
 Doctoral student Peng Xie came to The University of Texas at Dallas primarily to follow his mentor, Dr. Michael Q. Zhang, and his passion, computational biology.
Doctoral student Peng Xie came to The University of Texas at Dallas primarily to follow his mentor, Dr. Michael Q. Zhang, and his passion, computational biology.In the three years he has been on campus, Xie has exemplified the multitude of reasons that promising graduate students come to UT Dallas — opportunity, support and career advancement.
Xie is a member of the first cohort of the Eugene McDermott Graduate Fellows Program. In addition to a stipend, tuition and fees, and a research budget, the McDermott fellowship provides professional development and enrichment opportunities.
Scientist Strengthens Systems Biology Initiative
 Dr. Michael Zhang, a leading scientist in computational biology and genomic research, has joined UT Dallas as professor and Cecil H. and Ida Green Distinguished Chair of Systems Biology Science. Zhang’s chaired position in the School of Natural Sciences and Mathematics begins establishment of a new Center for Systems Biology. “In attracting a researcher of the stature of Dr. Zhang to our faculty, we expect to nucleate a major effort in genomics and computational biology that focuses on the genetic underpinnings of disease,” said Myron Salamon, dean of the School of Natural Sciences and Mathematics. Computational biology bridges the life sciences and quantitative sciences – mathematics, statistics and computer science – to understand living systems.
Dr. Michael Zhang, a leading scientist in computational biology and genomic research, has joined UT Dallas as professor and Cecil H. and Ida Green Distinguished Chair of Systems Biology Science. Zhang’s chaired position in the School of Natural Sciences and Mathematics begins establishment of a new Center for Systems Biology. “In attracting a researcher of the stature of Dr. Zhang to our faculty, we expect to nucleate a major effort in genomics and computational biology that focuses on the genetic underpinnings of disease,” said Myron Salamon, dean of the School of Natural Sciences and Mathematics. Computational biology bridges the life sciences and quantitative sciences – mathematics, statistics and computer science – to understand living systems.
Affiliations
Posdoctoral Fellow
1987/07–1990/12Courant Institute of Mathematics, NYU
GenomeFellow/Assistant/Associate/Full Professor/AdjunctFaculty
1991/01–2010/06Cold Spring Harbor Laboratory and Watson School of Biological Sciences
Adjunct Faculty
1991/01–2010/06Genetics/Physics/Math, SUNY / Stony Brook University
Adjunct Faculty
2005/07–2021/11School of Informatics Sciences/School of Medicine, Tsinghua University